You Are What You Eat: The Influence of Food Microbiomes on Human Microbiomes
What 2,500 Food Metagenomes Tell Us About Human Microbiomes
Subscribe to Genomely for the latest discoveries and in-depth analyses in your inbox
Thank you to our subscribers for your continued support and passion for science!
In recent years, exploring the microbiomes within our food has gained tremendous traction. While the connection between the human gut microbiome and overall health is widely acknowledged, less attention has been given to the microbes present in the food we consume. Yet, food represents one of the most direct and frequent ways we interact with our resident microbial communities. A groundbreaking study recently published dives deep into this relationship, shedding light on the immense and largely untapped diversity of microorganisms found in food and their role in influencing human health.
A recently published study in the journal Cell by Carlino et al. generated the curatedFoodMetagenomicData (cFMD) resource, a database of over 2,500 integrated and analyzed food metagenomes. The study goes beyond simply identifying microorganisms—it maps out how the microbes in our food impact the composition of our gut microbiome.
Here, we will cover the main findings from the study, explore the implications for food science and human health, and discuss the exciting new avenues for future research that this work opens up.
The Rise of Metagenomics in Food Microbiology
Microorganisms have always played a critical role in food production and preservation, from fermentation in dairy, meat, and plants, to spoilage and contamination. For centuries, humans have developed methods to harness beneficial microbes for food preservation and flavor enhancement, such as fermenting milk into cheese or using yeast to bake bread. However, much of the microbial diversity in food remained unexplored until the advent of metagenomics—a powerful tool that allows scientists to study the genetic material of entire microbial communities without needing to culture individual species.
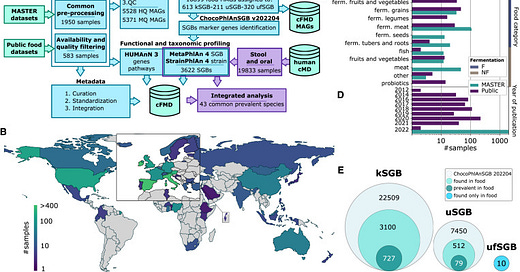
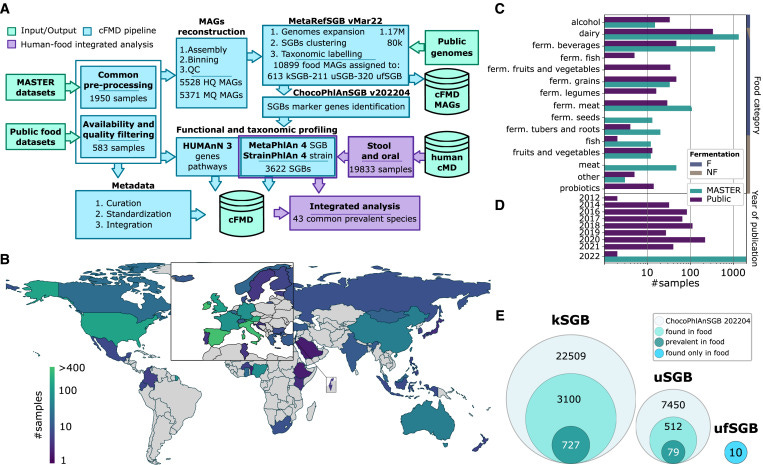
Previous studies have largely focused on isolated food categories or specific types of food microbiomes. However, this new research took a broader approach, integrating newly sequenced and publicly available food metagenomes to build a comprehensive resource. The result is a dataset that includes over 10,000 metagenome-assembled genomes (MAGs), providing the most extensive view of the prokaryotic and eukaryotic species present in our food.
The Diversity of Food Microbiomes
The cFMD resource revealed remarkable microbial diversity across different food types, uncovering over 1,000 species-level genome bins (SGBs) from both prokaryotic (bacteria and archaea) and eukaryotic (fungi and other microbes) organisms. Surprisingly, 320 of these species had never been described before. This finding highlights how little we know about the microbes living in and on our food.
One of the key discoveries of this study is that different food categories (e.g., dairy, fermented beverages, and meats) host distinct microbial communities. Foods like cheese, kefir, and sauerkraut, which undergo fermentation, naturally harbor rich microbial populations. Even within these categories, there is significant variability, with some foods containing highly specialized microbial communities. For example, fermented seeds, tubers, and raw milk showed particularly high levels of previously uncharacterized species.
By categorizing these food-associated species into distinct SGBs, the researchers created a foundational dataset that enables future exploration of how different food microbiomes interact with one another and with the human gut.
You Are (Somewhat) What You Eat: Food and the Human Microbiome
One of the most intriguing aspects of this research is the connection between food microbiomes and the human gut microbiome. It has long been understood that the food we eat influences the composition of our gut microbes, but the specific role of microbes transmitted from food to the gut has remained largely unexplored. This study addresses this gap by analyzing over 20,000 human metagenomes and food metagenomes in the cFMD resource.
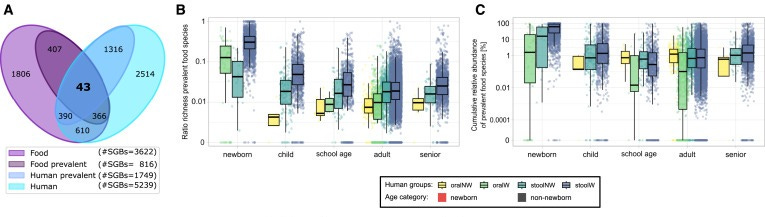
The researchers found that food microbes account for an average of 3% of the adult gut microbiome. While this may seem like a small percentage, it represents a significant overlap, especially considering the complexity of the human gut microbiome, which consists of thousands of microbial species. This overlap was even more pronounced in infants, where food-related species comprise a much more significant portion of the gut microbiome—up to 56%. This finding suggests that dietary microbes play a critical role in shaping the development of gut microbiome early in life.
Strain-Level Analysis: Tracing Microbial Transmission from Food to Gut
While metagenomics has enabled broadly charting the species and genera present in a microbial consortium, identifying strain-level presence (i.e., a particular lineage within a species) is more difficult. However, this study presents a compelling strain-level analysis, revealing potential instances of food-to-gut microbial transmission. For example, the study identified specific strains of Lacticaseibacillus paracasei—a bacterium commonly found in fermented dairy products—that appear to be transferred from food to the gut. The researchers also found that food-borne Limosilactobacillus mucosae and Streptococcus gallolyticus differ genetically from those found in the human gut, raising intriguing questions about how these strains may adapt or evolve in different environments.
This strain-level analysis provides crucial insights into how microbes might move between food and the human gut and how specific microbial strains from food may establish themselves as permanent or transient members of the gut microbiome.
cFMD is a Treasure Trove for Food Science
The cFMD resource opens up many exciting opportunities for future research and applications in food science, human health, and biotechnology. Here are a few potential uses:
1. Food Safety and Quality
Metagenomics can be used to monitor the microbial communities in food products to ensure their safety and quality. By identifying the specific microbial signatures associated with food spoilage, contamination, or disease, researchers can develop better methods for preventing foodborne illness.
2. Microbiome-Driven Food Authentication
One of the most exciting prospects is using food-specific microbial signatures for food traceability and authentication. For instance, certain microbial species were found to be highly specific to particular food categories or even geographic regions. This opens the door for developing microbial markers that can verify the authenticity of food products, particularly in industries like wine, cheese, and fermented foods, where terroir plays an important role.
3. Dietary Microbiome Interventions
Understanding how dietary microbes influence the gut microbiome can help develop targeted dietary interventions to improve gut health. For example, future research could investigate whether consuming specific fermented foods can promote beneficial strains of microbes in the human gut, potentially offering new ways to prevent or treat conditions like obesity, inflammatory bowel disease, or even mental health disorders.
4. Biotechnological Applications
The newly discovered microbial species in the cFMD resource also represent a vast untapped resource for biotechnology. These microbes could be harnessed for their unique enzymatic properties or fermentation capabilities, potentially leading to the development of new food products, probiotics, or industrial processes.
Challenges and Future Directions
While the cFMD resource is a monumental step forward in understanding the intersection of food and human microbiomes, there is still much work to be done. For one, detecting microbial transmission from food to humans is challenging. While this study identified potential cases of food-to-gut transmission, further research is needed to confirm these findings and explore the mechanisms behind this transmission. For example, it is still unclear whether microbes from food that enter the gut are merely transient or whether they can establish long-term colonization.
Additionally, the study highlights the need for more diverse sampling. The cFMD resource is heavily weighted toward certain food types (like dairy) and geographic regions (primarily Europe and North America). Expanding the database to include more foods from different cultures and regions worldwide would provide a more comprehensive picture of the global diversity of food microbiomes.
Conclusion: A New Frontier in Microbiome Research
This study represents a watershed moment in our understanding of food microbiomes and their impact on human health. By integrating thousands of food and human metagenomes, the researchers have created a resource that not only expands our knowledge of the microbial diversity in food but also provides crucial insights into how these microbes interact with our gut microbiomes.
The curatedFoodMetagenomicData (cFMD) resource is more than just a scientific achievement—it’s a starting point for a new era of food science. Whether it’s improving food safety, developing microbiome-based dietary interventions, or unlocking new biotechnological innovations, the possibilities are vast and exciting.
As research in this field continues to grow, one thing is certain: the microbes in our food are far more than passengers. They are active participants in our health, and understanding their role is key to unlocking the full potential of the microbiome revolution.